Presentations from the FSA Fellow and PhD Student
On this page
Skip the menu of subheadings on this page.Introduction
1. The FSA and COT have been reviewing New Approach Methodologies (NAMs) to scope the best scientific methodologies available to be used in the risk assessment of chemicals in foods and the environment and understand how these can be incorporated and accepted in a regulatory context.
2. In 2021, the FSA funded a computational toxicology postdoctoral fellow Dr Arthur de Carvalho e Silva at the University of Birmingham and a PhD Student Mr Alexander Kalian (London Interdisciplinary Doctoral Program-LIDo-TOX AI) at King’s College London.
3. The fellow and PhD student have been working alongside other government departments to understand how NAMs will improve indicative levels of safety in chemical risk assessment.
4. In addition, these new partnerships have helped with networking, research collaboration, training opportunities and furthering in this area. The fellowship and studentship also compliment the work set out in the UK Roadmap towards using new approach methodologies in chemical risk assessment.
5. The fellow and the PhD student have prepared a yearly review as outlined below and will present their progress to date to the COT Members.
Postdoctoral Fellow
Advancing in silico methods of assessing toxicological risk
Why and how are you associated with the FSA?
6. In 2021, the FSA elaborated a roadmap for the implementation of new approach methodologies (NAMs) for the risk assessment of potentially harmful substances in the UK regulatory landscape. One of the activities defined in this roadmap was funding an FSA postdoctoral fellow to actively work on advancing in silico methods for assessing toxicological risk, specifically focused on food-related chemicals, but remaining open to work on other classes of chemicals relevant to the FSA’s risk assessments. In this context, I was recruited as a computational toxicology fellow and awarded a 4-year fellowship funded by the FSA, whilst supervised by a team of academic and applied NAM experts. The supervisory team is composed of Prof. Mark Viant and Prof. John Colbourne (University of Birmingham), Dr. George Loizou (HSE Science and Research Centre), and Dr. Olivia Osborne, Ms. Claire Potter, and Dr. David Gott (FSA).
Broad overview of FSA fellowship and its aims
7. The programme of work in the fellowship consists of (i) scoping the FSA’s problem space in chemical risk assessment and mapping this to our computational NAMs solution space, thereby aiding the FSA to develop a strategy for the utilisation of NAMs (months 1-24); (ii) ensuring that the FSA is trained in the use of computational NAMs by delivering training courses, including an introduction to existing and emerging NAM technologies, and topics selected from the FSA’s NAM strategy (months 1-48); (iii) developing and evaluating confidence in a new hazard assessment workflow that integrates in vitro omics toxicity data, benchmark dose modelling and PBPK modelling to serve as the basis for quantitative risk assessment for human health, i.e. towards generating human health-based safety thresholds for FSA and other regulators (months 1-36); and (iv) developing and delivering a second case study that fortifies the community-wide acceptance of 21st century methods in risk assessments, to accelerate the successful application of NAMs within the FSA (months 25-48).
Progress with the first case study
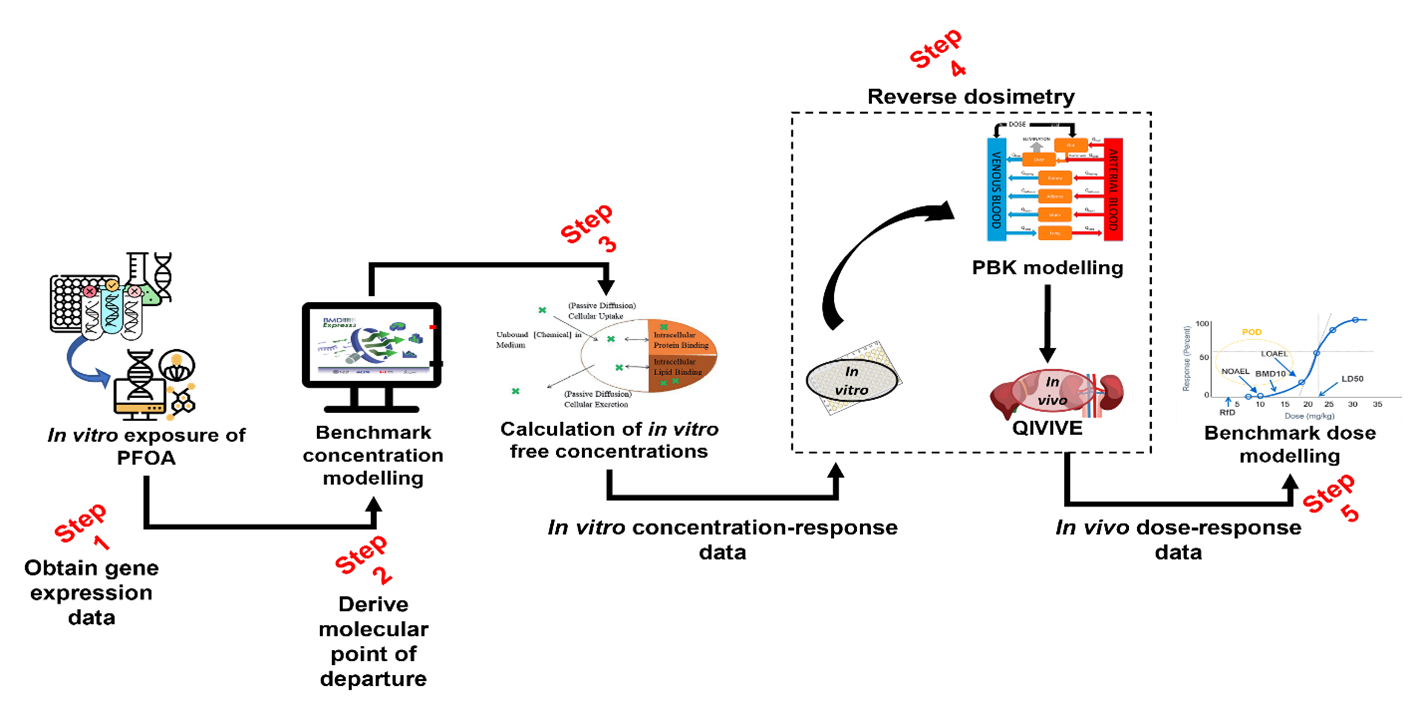
8. The workflow implemented and tested is described in Figure 1. Through its five steps, it seeks to utilise multiple NAM approaches, ultimately to generate a human-health based point of departure for risk assessment. The NAMs being employed include:
NAMs in relation to the type of testing platform - in vitro hepatic microtissues.
NAMs in relation to the type of data/read-outs - transcriptomics data, providing an untargeted measurement of extensive gene expression.
NAMs in relation to data analysis - PBPK modelling.
9. The systematic integration of various data generation and modelling approaches has required a broad range of expertise, with steps 1-2 predominantly guided by the University of Birmingham team and steps 3-5 by the HSE Science & Research Centre.
10. To date, two case studies were conducted. The first case study focused on the plasticiser di-2-ethylhexyl terephthalate (DEHTP). The main objective was to derive a health-based guidance value mainly focusing on steps 3-5 of the workflow (Figure 1). Concentration-response data obtained from ToxCast, via the Chemicals Dashboard (US EPA), was used. The second case study had as chemical of choice a perfluorinated substance, perfluorooctanoic acid (PFOA). The main objective was to integrate our insilico workflow with transcriptomics data to derive a health-based guidance value for PFOA that could be compared with that previously published by the European Food Safety Authority (EFSA). Transcriptomics data published by Health Canada (Rowen-Carrol et al 2021) was used as a data source from in vitro exposures of human liver microtissues to PFOA.
Figure 1. In silico workflow to generate a human health-based point of departure for risk assessment.
Progress with papers and conferences
11. The DEHTP and PFOA case studies are both intended for publication. It will report the utility of the workflow and the integration of the calculation of in vitro free concentrations as an important step towards deriving a health-based guidance value. The second paper is written and being revised by the co-authors. Our recent work on PFOA was presented on several occasions. To list a few, PARC Science Day (poster presentation), NURA Dynamic Discussions (oral presentation, online), HSE’s workshop (oral presentation, online), EFSA’s workshop (oral presentation, online), EUROTOX 2023 (poster presentation), ASPIS Open Symposium 2023 (poster presentation). Our second case study project was submitted as a nomination to the Lush Prize under the Young Researcher category and has been one of the five projects awarded in 2022. The third case study is now under consideration by the supervisory team, which is keen to work with tropane alkaloids as this class of substances is of high interest of the FSA.
PhD Student
TOX-AI : Digitalising toxicological databases using Artificial Intelligence and in silico tools for food safety
Association with the FSA
12. The Food Standards Agency (FSA) is jointly funding this PhD project, alongside the Biotechnology and Biological Sciences Research Council (BBSRC), as an iCASE (industrial CASE) project under the LIDo (London Interdisciplinary Doctoral programme) consortium, with the PhD project itself based at King’s College London, while the FSA is named as a partner. Dr Olivia Osborne, Dr David Gott and Ms Claire Potter, of the FSA, are all formally named as part of the supervisory team, in addition to other supervisors. This project shall contribute to the FSA’s interests, in developing innovative Artificial Intelligence (AI) based New Approach Methodologies (NAMs) for next-generation chemical risk assessment of molecules found in food and drink, to assist in improving consumer safety while simultaneously reducing reliance on animal testing. It is intended that, in addition to numerous publications of interest, a corresponding database of high-quality predictions for certain food and drink molecules shall be released for the FSA’s use, alongside an open-source software tool. As the PhD student, I am also required to complete a placement within the FSA, as part of this PhD project.
Broad Overview of PhD
13. The primary aim of this PhD project is to develop novel Quantitative Structure-Activity Relationship (QSAR) models, using innovative AI, which may reliably predict toxicological properties of molecules found in food drink, over a diverse range of endpoints of interest. Having already developed QSAR models of mutagenicity, using deep learning, the research is now progressing into predicting for other more challenging endpoints, such as neurotoxicity, developmental and reproductive toxicity, endocrine disruption and others, with predictions concerning both toxicokinetics and toxicodynamics. Particular research efforts are also being focused on Brominated Flame Retardants (BFRs) as a case study, in order to highlight BFRs of particular concern or which alternatively may simply require more extensive data collection. In addition to progress made in terms of endpoints, progress is also aimed with regards to the types of AI used, especially via more advanced deep learning architectures in the Graph Neural Network (GNN) space. It is ultimately intended that the final models produced by this project will be as accurate as possible in their predictions, while also providing unique insights into toxicological space, compared to existing models in literature. It is furthermore aimed that the final developed QSAR models will be explainable and easily interpretable, via use of explainable AI (XAI), along with reliable quantifications of uncertainty on particular predictions. The final QSAR models shall also be made available via an open-source software tool, accompanied by a database of associated predictions for certain molecules. The methods, results and other materials are being developed in close collaboration with the FSA, with a placement within the FSA due to take place, while all materials are to be published in open-access publications and/or presented at relevant conferences.
Main Work Up to this Point
14. The main work up to the present is composed of three parts: (1) Exploration of dimensionality reduction algorithms, for powering QSAR models of mutagenicity, constructed of simple feed-forward Deep Neural Networks (DNNs) (Kalian et al., 2023, Kalian et al., 2023); (2) Development of Graph Convolutional Networks (GCNs) to improve mutagenicity predictions, via graph classification of molecules, while also allowing for mining of structural alerts (SAs); (3) Development of GNNs for node classification of molecules, in order to predict toxicological properties of BFRs, starting with acute toxicity and comparing to predictions from the Toxicity Estimation Software Tool (TEST) of the US EPA.
Progress with papers and conferences
15. Published 2 papers: Exploring Dimensionality Reduction Techniques for Deep Learning Driven QSAR Models of Mutagenicity (Kalian et al., 2023) and Improving accuracy scores of neural network driven QSAR models of mutagenicity (Kalian et al., 2023).
16. I gave a talk at ESCAPE 33 (The 33rd European Symposium on Computer-Aided Process Engineering) in Athens, Greece, in April 2023. My talk (and conference proceedings paper) was titled: "Improving accuracy scores of neural network driven QSAR models of mutagenicity".
17. I also presented a poster at WC12 (The 12th World Congress on Alternatives and Animal Use in the Life Sciences 2023) in Niagara Falls, Canada, in August 2023. My poster was titled: "Dimensionality Reduction Algorithms for Powering Deep Learning Ensemble QSARs of Mutagenicity".
18. Paper in progress (creating manuscript): “Combining Graph and Language Based Explainable Artificial Intelligence, for Mining Structural Alerts of Mutagenicity”.
19. Conference abstract in progress: Deciding between EUROTOX or SOT in 2024, to present work on node classification of BFRs.
Questions for the Committee
Following the presentations, Members will be invited to ask questions.
References
Kalian, A.D., Benfenati, E., Osborne, O.J., Gott, D., Potter, C., Dorne, J.L.C., Guo, M. and Hogstrand, C., 2023. Exploring Dimensionality Reduction Techniques for Deep Learning Driven QSAR Models of Mutagenicity. Toxics, 11(7), p.572. https://doi.org/10.3390/toxics11070572
Kalian, A.D., Benfenati, E., Osborne, O.J., Dorne, J.L.C., Gott, D., Potter, C., Guo, M. and Hogstrand, C., 2023. Improving accuracy scores of neural network driven QSAR models of mutagenicity. In Computer Aided Chemical Engineering (Vol. 52, pp. 2717-2722). Elsevier. https://doi.org/10.1016/B978-0-443-15274-0.50432-7
Rowan-Carroll A, Reardon A, Leingartner K, Gagné R, Williams A, Meier MJ, et al. Toxicological Sciences. 2021;181(2):199-214. https://doi.org/10.1093/toxsci/kfab039